The Skinniest Data Pipeline You Have Ever Seen
Here at Pollen, we recently released a dataset containing billions of rows of climate data hosted in Snowflake. When I began the project, I was laboring under the illusion of infinite time and, perhaps unwisely, decided it was a good idea to add an additional challenge: construct the data pipeline with as few frameworks and dependencies as possible, just to see if I could do it.
Well, I did it.
My motivation? I’ve been building software for 25 years. In that 25 years, computing power has increased exponentially. But, you know what hasn’t improved exponentially? The preceived wall time of actually executing software to create a discrete unit of business value. That feels like it’s about the same as it’s always been. It takes about as long to render a web page as it always has. It takes about as long to load a file as it always has. It costs about the same to run a cluster as it always has. Or, at least it feels that way.
I had this emotional feeling that these things take so long, still, because we use so many frameworks and layers of abstraction that we’re drowning in their overhead and don’t even know it. To me, our software stacks feel like that one person’s house on your block at Christmas, where they switched from incandescent Christmas lights to LED Christmas lights five years ago and, instead of reveling in the glory of saving 90% of their electricity usage, they instead added 10x more lights. We’ve added 10x more lights to our software stacks, just because we can. So, I wanted to test that theory by creating the skinniest data pipeline I could conceive of, just to see what it would look like.
I still wanted to keep conceptual best practices, though
In building this data pipeline, I still wanted to incorporate best practices I’ve learned over the years, though. Yeah, I could have built a toy pipeline that runs as one, giant, in-process Python script on my laptop, but that’s not really a good comparison. I wanted my pipeline to be production quality. Which means it exhibits the following qualities:
- Asychronous steps tied together with a message passing paradigm
- ...broken up into descrete steps that can fail and retry independently
- ...that gets source data "in the door" as soon as possible and saves it in raw form on infrastructure we control
- ...that catches unanticipated errors cleanly and in a way that can be debugged, fixed, and retried
- ...that is reentrant and idempotent
- ...that scales seamlessly up to thousands of nodes in the cloud AND down to one node on my laptop
- ...that can split CPU-bound and IO-bound loads onto different hardware, if I discover such a thing is a problem
- ...that's containerized such that you can run EXACTLY the same image on your laptop as on production
At a "business logic" level, I needed 7 steps
Logically speaking, my pipeline consisted of 7 steps:
- Given high-level search criteria, query the Earth Systems Grid Federation API to find logical datasets and the URL(s) of their netCDF data files. Package up what we find into discrete, high-level units of work (one per logical dataset) and publish those units of work onto a queue to be done in parallel.
- ...download the actual source file from whatever university lab it's stored on and upload it again to a predictable place in S3
- ...read the metadata of the netCDF file itself to find auxiliary information that happens to be stored in yet more, different files on other academic servers (I called this "stitching")
- ...store all the dataset's stitched-together metadata in one, coherent "envelope" that I send down the pipeline and also store in a table in Snowflake for easy reference later
- ...fork parallel sub-units of work to transform each of the dataset's constituent files (there might be dozens or even hundreds per dataset) from netCDF into a tabular csv format and store that csv in S3
- ...copy/import the csv from S3 into a staging table in Snowflake and then merge/upsert from that staging table into a gigantic, target fact table
- ...store some more metadata in Snowflake
I wanted each of those steps to happen independently (but in serial), and I wanted each of them to serve as their own kind of “checkpoint” on the process. In other words, if step 5 failed, I only wanted to have to fix the problem in step 5 and then re-run step 5, without having to first roll all the way back to step 1. And I really, really wanted step 1 and step 2 (the steps with the most external dependencies) to be as discrete as possble, so that this process was entirely under my control as close as possible to the “front door”. This is important because the academic servers that I’m having to rely on aren’t the most powerful nor reliable in the world.
So here's what the pipeline ended up looking like
I ended up building a pipeline that had Amazon SQS at its core and had plain, old, Python classes running inside a Docker container inside an Amazon ECS cluster pulling things off the SQS queue. The Python code pulled a task/envelope off the queue, added some value, and then published the next step back out to the same SQS queue. It’s about 2,500 lines of code and easily 80% of that 2,500 lines is pure, unadulterated business logic specific to my domain and the remaining 20% is plumbing, infrastructure, or file handling code.
The components look like this:
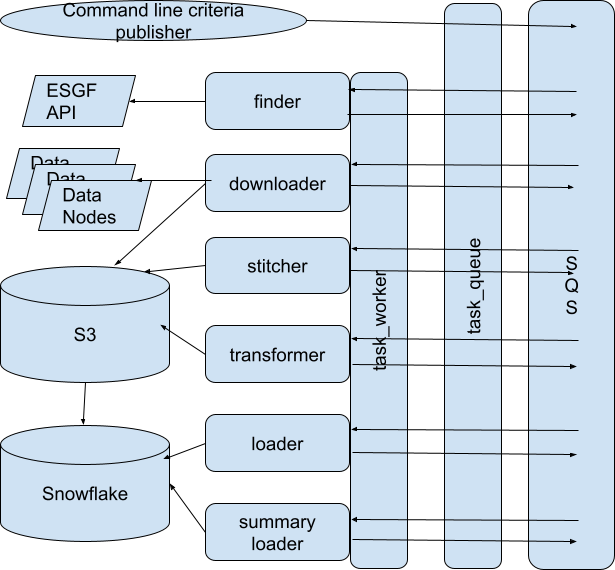
There is absolutely no third-party framework code; the “task worker” abstraction is, essentially, the only “framework” involved and it is 93 lines of code, including empty lines, with a main method.
The “task queue” is simply a queue-like abstraction over SQS, such that I could swap out SQS for, say, Redis or RabbitMQ or even an in-memory data structure. In fact, I also have a “blob store” abstraction over S3 and a “data store” abstraction over Snowflake. Each of these abstraction layers is less than 150 lines of code. Incidentally, I consider creating such abstraction layers over third party serivces to be a best practice. It’s a hundred-or-so lines of code well spent.
The task worker takes a JSON envelope off the queue, reads a “task_type” property that it assumes will be there, and has a big switch statement that instantiates a different task class (i.e. “downloader” vs “transformer”) depending on the task type, and then blindly hands off the rest of the JSON envelope to that class. The class is expected to enqueue the next step in the chain itself if it’s successful and it’s expected to throw an exception if it’s not. If the task class throws an exception, the worker “framework” increments a number of retries in the envelope and if that number is less than a configurable number, it re-enqueues the same envelope at the back of the queue. If the retries exceed the limit, it places the envelope on a different dead letter queue.
That’s it.
In fact, here is all the “framework” code so you can see for yourself. The “self.tq” is a reference to my task_queue abstraction, which is, itself, just a thin abstraction over the boto3 SQS library:
def start(self):
while True:
print("polling...")
self.tq.poll(self.on_message)
print("done polling")
def on_message(self, message):
self.clear_tmp_directory()
task_type = message['task_type']
attributes = message['attributes']
retries = attributes.get('retries')
if retries is None:
attributes['retries'] = 0
retries = 0
try:
if task_type == TaskQueue.TASK_FIND_DOCUMENTS:
CMIP6FileFinder(self.tq).poll(attributes)
elif task_type == TaskQueue.TASK_DOWNLOAD_DOCUMENT:
CMIP6FileDownloader(self.tq).download_and_enqueue(attributes)
elif task_type == TaskQueue.TASK_STITCH_DOCUMENT:
CMIP6FileStitcher(self.tq).stitch_document(attributes)
elif task_type == TaskQueue.TASK_TRANSFORM_FILE:
CMIP6FileTransformer(self.tq).transform_and_stage(attributes)
elif task_type == TaskQueue.TASK_LOAD_FILE:
CMIP6FileLoader(self.tq).load_file(attributes)
elif task_type == TaskQueue.TASK_LOAD_SUMMARY:
DatasetSummaryLoader().load_all(attributes)
else:
raise Exception(f"Unrecognized task queue message: {message}")
except Exception as e:
exception_message = str(e)
exception_type = str(type(e))
import traceback
exception_trace = traceback.format_tb(e.__traceback__)
print(f"Caught exception processing message {task_type} num retries {retries} with message {exception_type}: {exception_message} and trace {exception_trace}")
attributes['exception'] = {
'message': exception_message,
'type': exception_type,
'trace': exception_trace
}
retries += 1
attributes['retries'] = retries
if retries > self.max_retries:
print(f"Sending message to DLQ")
self.tq.publish_to_dead_letter_queue(task_type, attributes)
else:
print(f"Reenqueueing message")
self.tq.publish_to_regular_queue(task_type, attributes)
The pipeline performed beautifully
With this framework, I’ve transformed and then loaded several hundred billion rows of data into Snowflake using one ECS cluster with between one and five pretty small (1 vCPU) nodes.
The entire pipeline is CPU-bound on the transform step, which has nothing to do with my framework and everything to do with looping through netCDF files, which is a domain-specific problem. And this CPU-bound transform step can run “embarassingly parallel”, as in for every node I add to my cluster, I get an almost exactly linear improvement in throughput, which is how it should be.
The only dependencies this entire thing has are:
- The AWS Python API (boto3)
- The Snowflake Python library
- A netCDF reading library (it's a special, scientific, binary format)
- The "requests" Python library
- The "shapely" Python library (for properly formatting WKT geographic polygons to store in Snowflake)
- ...and the transitive dependencies for all of the above
That would have led to a tiny, tiny Docker image, except for one thing…
My biggest source of dependencies was a surprise
I really, really, really wanted to avoid having a dependency on numpy and pandas, especially. Once you bring in those two, then you bring in a world of hurt around a huge dependency graph that really wants to compile native C libraries at every turn. And two surprising dependencies foiled me.
First, the netCDF library foiled me. Even though I didn’t use any of the functionality, it nonetheless has a function or two that allowed you to inhale the file’s contents and present them as a numpy dataframe. I spent days trying to get a Docker image to compile from an “alpine” base and finally succeeded, until…
…the Snowflake driver imported a different set of numpy-like dependencies, but this time with different and incompatible versioning. I spent many more days wrestling with this, stubbornly insisting on building everything up from the tiny alpine Docker image before I finally gave up, and just based my image on the HUGE “jupyter/datascience-notebook” image instead, which already has all these dependencies built in. This was a depressing failure, but I’d already lost enough days of my life.
I spent, easily, 10% of my total development time just getting all the libraries to compile on a fresh Docker image. By comparison, I spent probably 0.01% of my development time on my “framework” code and the remaining 89.99% of time actually manipulating and validating my data transformations (i.e. the domain-specific value add that is the point of this project). Those ratios aren’t too bad, and, in fact, the big chunk of time compiling dependencies actually validates my instinct that minimizing third party dependencies and frameworks can be a worthy goal, sometimes.
This approach was cheap and it was fast
Gratifyingly, even with this much-bigger-than-should-be-necessary Docker image, I was still able to run this entire pipeline on an absolutely tiny Amazon ECS cluster. One cluster with one service with one task definition, using 1 vCPU. That’s. It. With this cluster, I transformed and loaded billions upon billions of rows of data. This was very satisfying. And it was also very, very cheap.
My total Amazon bill through this experience was about $500. And, really, it could have been less than 1/3 of that, except, I had to reload the same giant set of data several times. Why? I had to fix several of my own mistakes and misunderstandings about the data, itself, as they came up. Nothing wrong with the infrastructure, just something wrong with my brain. Luckily, I’d written the code to be aggressively idempotent.
For comparison, if we use the top rack rate on the Stitch Data pricing page, which tops out at 300 million rows and $1,250/month and we extrapolate it to the ~300 billion rows I ended up loading, it would have cost me $1.25 million.
I’d say I got a bargain.
None of this is auto-documented, though
If you use a framework like Dagster or dbt Cloud, then your data pipeline gets documented automatically with pretty pictures like this (source: Dagster)
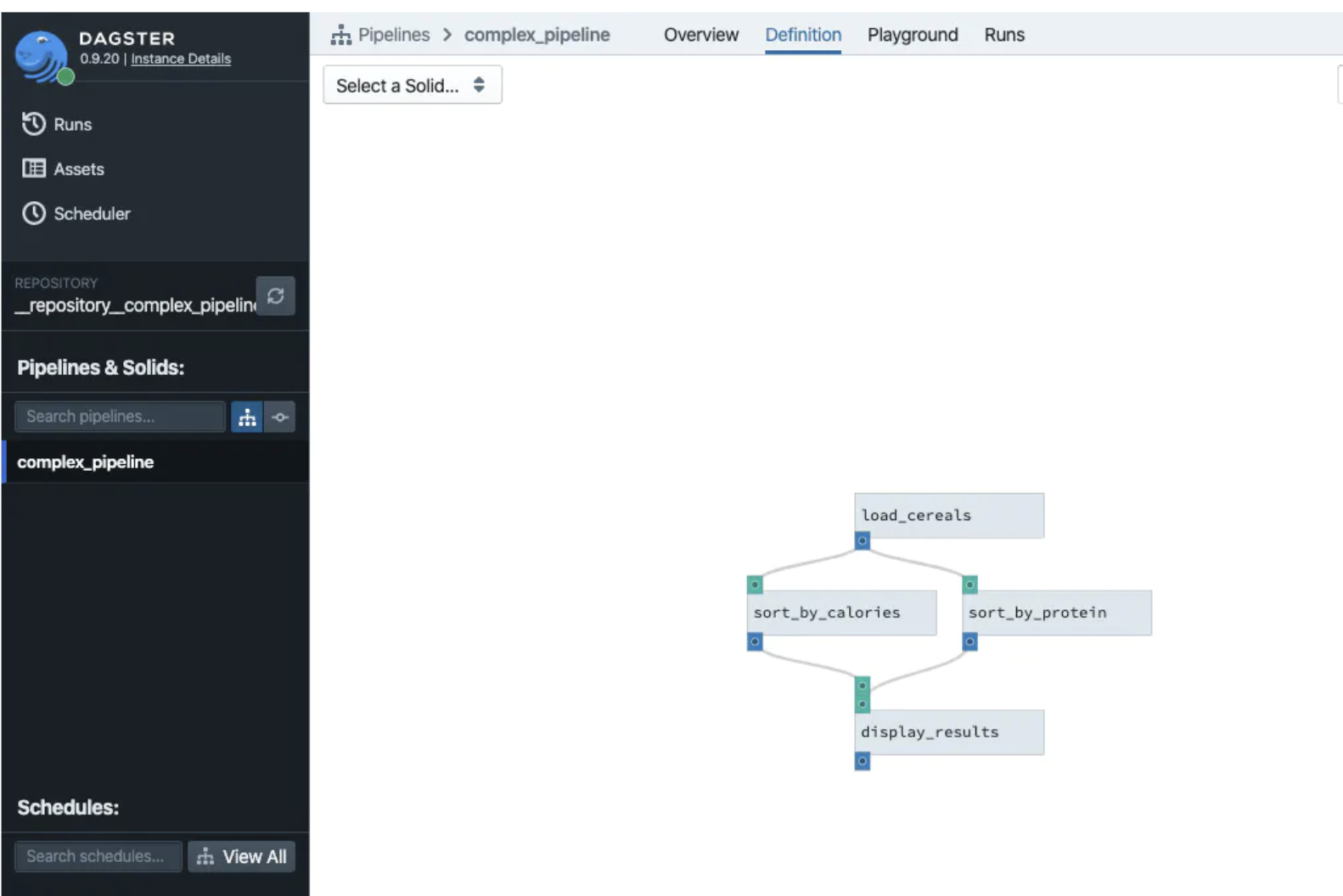
It’s tempting to neglect documentation, but you will absolutely regret it, eventually, when your company becomes a smashing success and is growing like crazy and you need to hire more people to scale. Or even when you, yourself, have to come back to this pipeline to modify something six months from now. For my purposes, this time, it was sufficient to take 15 minutes to draw my pretty drawing above in Google Docs, but most data pipelines need more documentation than that.
None of this is very observable
These three screenshots were my only window into what this pipeline was doing:
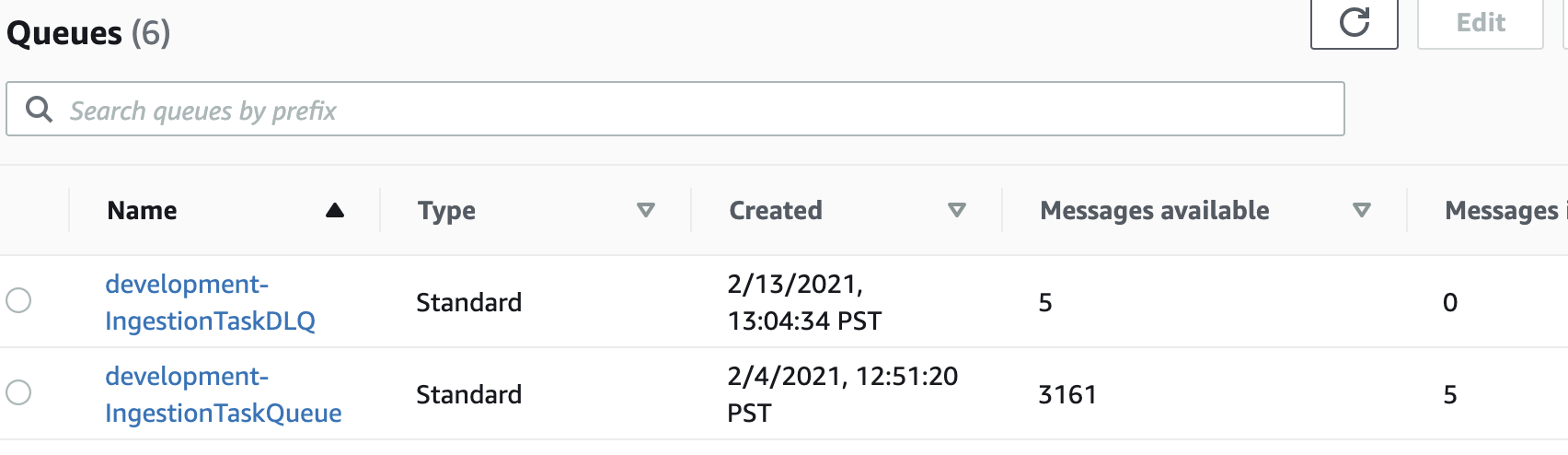
The SQS queue depth
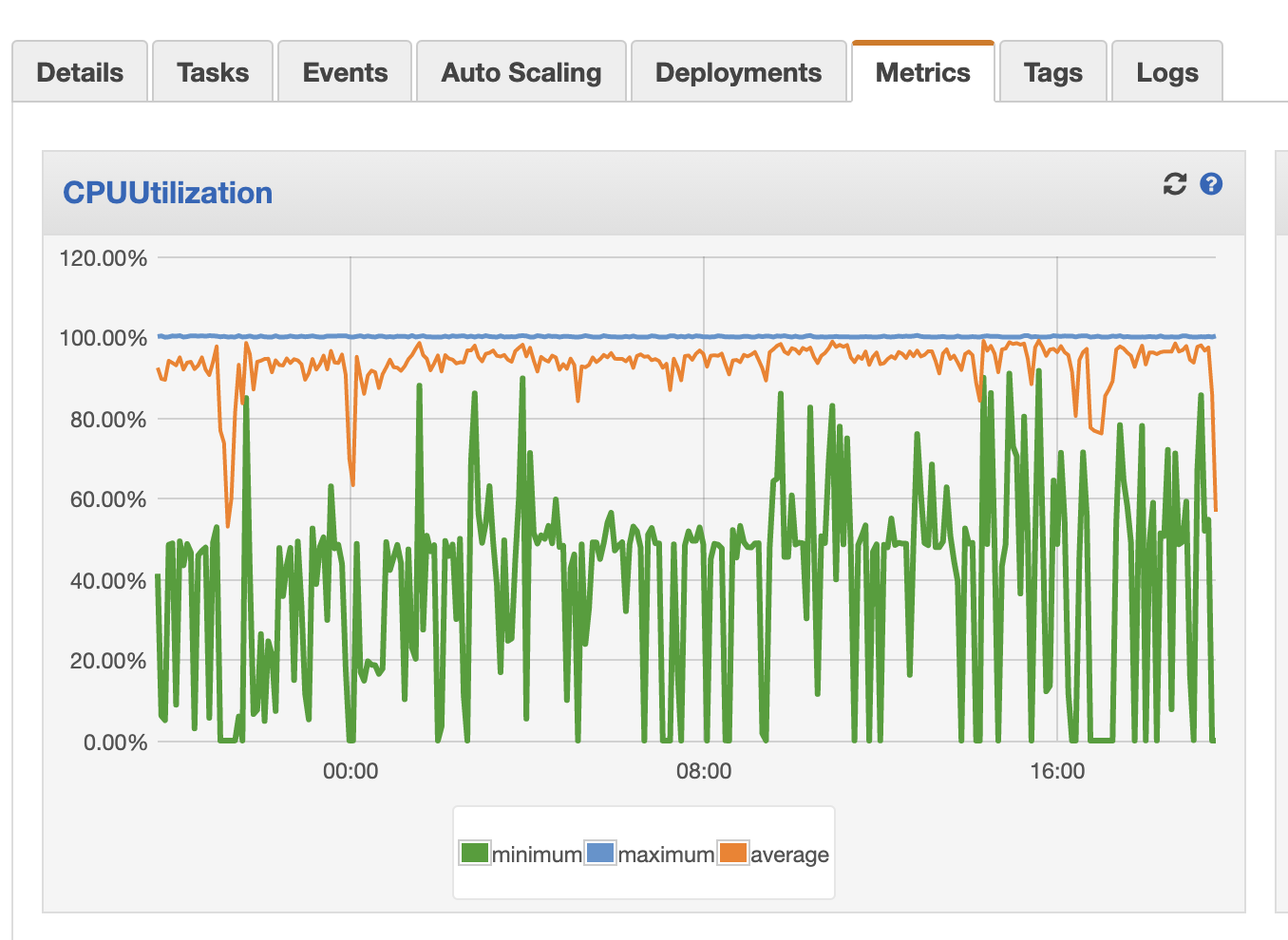
The ECS service CPU usage
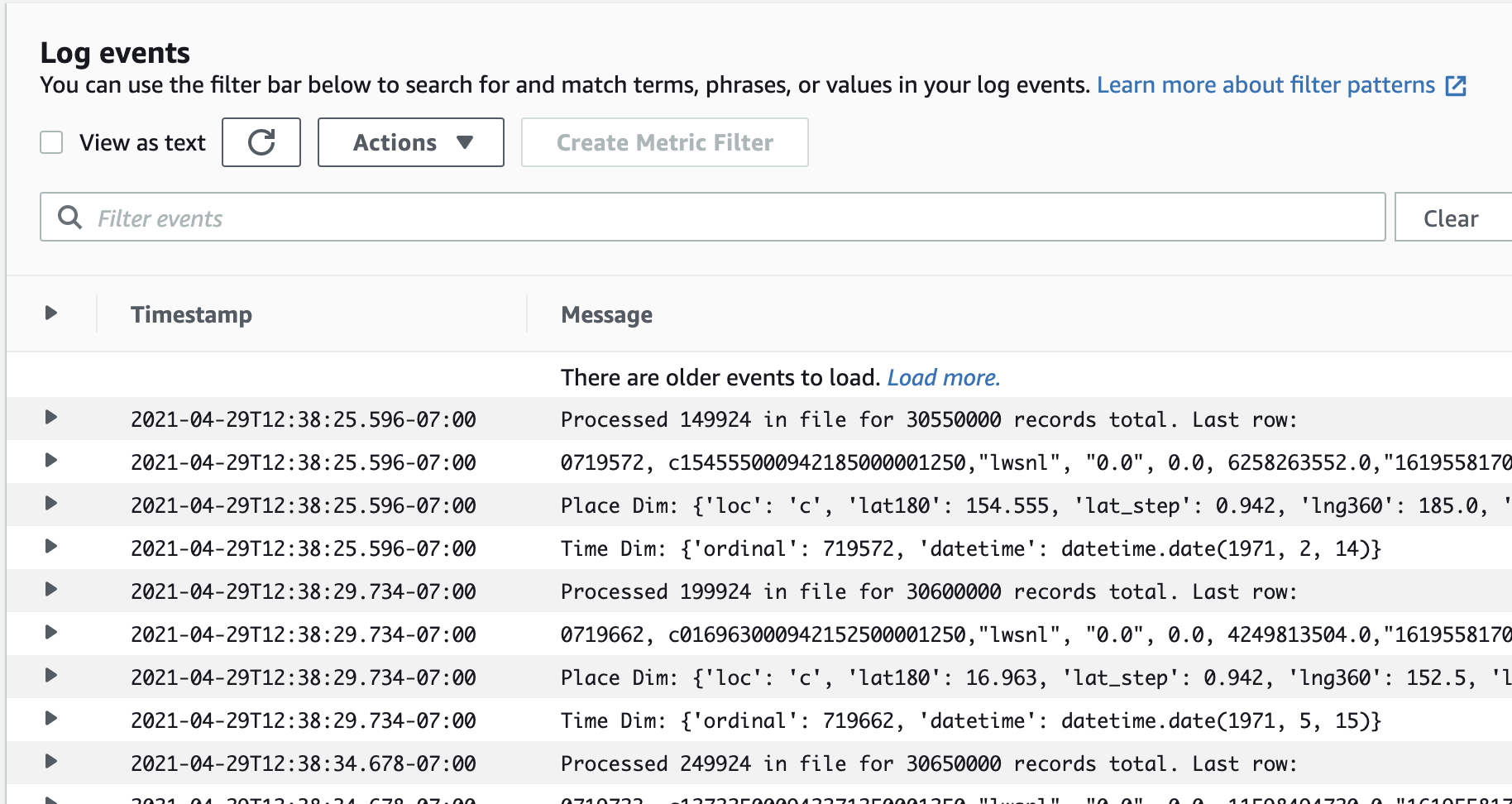
The ECS task CloudWatch logs
That’s…really not acceptable if this pipeline needs to see a lot of daily, variable action. It happened to be the case that my data pretty much gets converted once and doesn’t have to be touched very much afterward, so the lack of observability wasn’t too much of a problem. But, most data pipelines need much more observability than that.
The biggest problem, though, was split/join
My pipeline had two places where it logically split and one place where it logically joins. The first split is between the “finder” and the “downloader”. The finder gets a message like “find me all datasets that simulate temperature for these four climate scenarios and these six models” and then it will end up finding between zero and several dozen datasets.
The first split is when the finder enqueues a separate “download” task for each of these logical datasets. These datasets are the smallest, atomic “business unit of value”. One dataset might logically be “the precipitation projections from run r1f1p1i1 of the Community Earth Systems Model based on a worst case carbon scenario.” The actual data in that dataset spans several different files (sometimes a very large number of files) but, logically-speaking, users only ever deal with this dataset in its totality, as one logical unit.
Nonetheless, from a physical processing perspective, every one of the files in a given dataset can be independently converted to the CSV format we need. And that conversion is CPU-bound. So, converting all the files in a dataset is the kind of “embarassingly parallel” computing problem we all love to see. That’s where the second split comes from: between the “stitcher” that’s tracking down all the files, and the “transformer” that’s converting each of them to CSV in parallel.
Now, if I never join the pipeline together again after the transform step, it’s not the end of the world. The data loads into the Snowflake fact table in such a way that it all happens to work out in the end, even if I’m loading the contents of each file separately, in parallel. And then, after that load, I update a dataset-level metadata row in a table to say “this is basically what the dataset looked like”. If there are N files for one dataset, then I update that same metadata row N times with exactly the same data, once each time a file finishes loading. This is annoying and inelegant and potentially bug-prone, and it would cease to be a problem if only I could join my unit of work together again after the transform step.
The problem? Joining together a previously-split unit of work in a distributed pipeline is HARD to get right. There are all kinds of edge cases where you can go wrong. And most data engineering / ETL frameworks solve this problem for you. But… I wasn’t using a framework because I was being stubborn. And, so, I just suffered. This lack of “join” functionality was probably the biggest downside.
Conclusion: frameworks are good, but should earn their keep
This exercise was useful in that it highlighted the (often hidden) cost of using frameworks. If I’d had to bring in a lot more dependencies, then my compute cost would have been larger, my devops time would have been larger and more frustrating, and my cognitive overhead from mapping my problem onto the way the framework likes to think of things would have been higher. With this exercie, I know exactly what the smallest, cheapest, lowest-cognitive-load alternative is. I’ve got a baseline.
However. I should have used a framework anyway.
My specific use case happened to mostly dodge some thorny issues around documentation, observability, and hard things like split/join. And it happens to be the case that most of my pipeline’s processing happens only once, with just sporradic updates afterward. But, most data pipelines can’t dodge those issues, and so they need a framework to deal with them.
That said, now that I know the baseline cost of rolling my own, I insist that said framework earn its keep by adding value. And, I even know the areas where I’d expect it to earn its value the most: documentation, observability, and hard functionality like split/join. So, I’ve got my evaluation criteria already written for me. And I have an idea of how much it should cost.
2023 © Pollen Analytics LLC. ALL Rights Reserved.



